|
|
Amino acid description
- Absolute configuration at the alpha position
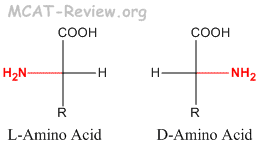
- L and D is different from R and S. L is not always S, and D is not always R.
- If the priority of NH2 > COOH > R, then L=S and D=R. For example, L-Alanine = S-Alanine.
- If the priority of NH2 > R > COOH, then L=R, and D=S. For example, L-Cysteine = R-Cysteine.
- L-amino acids are the more common in nature, and are the type found in proteins. D-amino acids are less common in nature, and are never found in proteins.
- Amino acids as dipolar ions classification
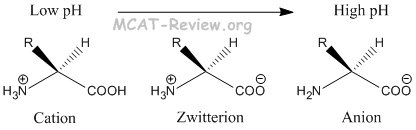
- At low pH, amino acids exist in the cationic form.
- At high pH, amino acids exist in the anionic form.
- At pH = pI, amino acids exist in the zwitterion form, which is overall neutral.
- Classification
- Acidic or basic
- If the R group contains carboxylic acid, then it's an acidic amino acid. There are two acidic amino acids: aspartic acid and glutamic acid.
- If the R group contains an amine group, then it's a basic amino acid. There are three basic amino acids: lysine, arginine, and histidine.
- Hydrophobic or hydrophilic
- Hydrophobic: If the R group doesn't contain any of the stuff below.
- Hydrophilic: If the R group contains acids, bases, amines or alcohols.
Amino acid reactions
- Sulfur linkage for cysteine and cystine
- Cysteine = side chain with the thiol group
- cystine = 2 cysteines forming a disulfide bond
- Peptide linkage
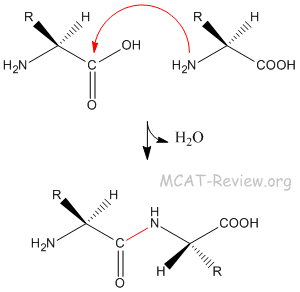
- Peptide bond = amide bond.
- The peptide bond is formed by the amine group attacking the carbonyl carbon.
- Hydrolysis
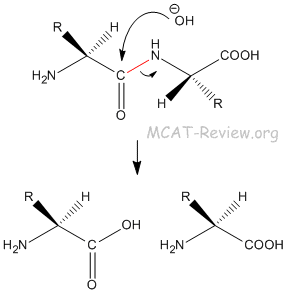
- The peptide bond is very difficult to hydrolyze. It requires a strong base, or a biological enzyme.
Protein structure
- Primary structure of proteins
- Primary structure = sequence.
- The primary structure of proteins is read from the N-terminus to the C-terminus.
- Secondary structure of proteins
- Secondary structure = repetitive motifs formed by backbone interactions.
- Backbone interactions = hydrogen bonding between the NH and C=O
- The two most common secondary structures are α helices and β pleated sheets.
- The α helix is right-handed, with the R groups sticking outward.
- In β sheets, R groups stick out above and below the sheet.
- Tertiary structure
- 3D structure of proteins
- Caused electrostatic side chain - side chain interactions
- Quaternary structure
- Separate chains/subunits joining together
- Caused by covalent disulfide bonding of cysteine side chains
Protein conformational stability
- Folding = chain -> 3D structure
- Many proteins fold spontaneously, some require the assistance of chaperone proteins
- Denaturing = loss of the native 3D structure such that it no longer function
- Extreme heat, salt concentration, or pH denatures proteins
Separation techniques
- Isoelectric point
- pH at which the molecule is neutral
- Acidic amino acids and proteins with lots of acidic side chains have have a lower isoelectric point
- Basic amino acids and proteins with lots of basic side chains have a higher isoelectric point
- Electrophoresis
- Protein is charged
- An electric field forces protein to travel through a gel
- Larger charge = more electrical force = travels faster
- Smaller protein = squeezes through easier = travels faster
- Structure plays no role because SDS is usually added to denature the protein
Non-enzymatic protein function
- Binding: the active site binds the substrate
- Stronger binding = lower Kd value
- Stronger binding doesn't necessarily mean a more efficient enzyme (if it binds the substrate and not let go, then it can't catalyze a new substrate)
- Stronger binding = better antibody
- Immune system
- Antibody = proteins that bind antigen on pathogens, which promote their destruction by the immune system
- Antigen = proteins expressed by pathogens. They can either directly bind to antibodies, or be presented by antigen presenting cells (such as macrophages and dendritic cells)
- Complement = proteins that punch holes in cells to be destroyed by the immune system
- Motors: uses ATP/GTP as energy to create motion
- Flagella (bacteria, sperm)
- ATP synthase (mitochondria, chloroplasts)
- Motile cilia (trachea)
- Myosin (muscle)
- Kinesin/Dynein (intracellular transport)
- Actin polymerization (Listeria)
Old topic, no longer tested
- synthesis of amino acids
- Strecker synthesis
- starting material: R-aldehyde
- reagents: cyanide (KCN), ammonium (NH4Cl)
- product: amino acid with the -R group originally on the aldehyde
- Gabriel synthesis
- starting material: R-halide
- reagents: 1. phthalimide, 2. NH2-NH2
- product: amino acid with the -R group originally on the halide
|
|
