|
|
Mendelian concepts
- Phenotype and genotype
- Phenotype: what is observed. For example, height, color, whether the organism exhibits a trait.
- Genotype: the genetic make up. For example, homozygous dominant (TT), heterozygous (Tt), homozygous recessive (tt).
- Gene: a gene is a stretch of DNA that codes for a trait. In molecular biology, the gene codes for a protein, which acts to bring about a trait.
- Locus: location (of a gene) on a chromosome.
- Allele: single and multiple
- An allele is a variant of a gene. A gene may have a number of alleles. All alleles of the same gene exist at the same locus.
- A cell holds 2 alleles of each gene. One allele from mom, one allele from dad.
- When a gene has only 2 alleles, then that's the simple case we're used to seeing. For example, the trait for height in peas is governed by T and t. TT and Tt gives tall plants, and tt gives short ones.
- When a gene has more than 2 alleles, then that's called multiple alleles. For example, blood type is governed 3 alleles: IA IB and i. Because a cell can only hold 2 of these alleles, the different combinations an individual can have are:
| Genotype | Blood type (phenotype) |
| IAIA or IAi |
A |
| IBIB or IBi |
B |
| IAIB |
AB |
| ii |
O |
- Homo- and heterozygosity
- Homozygous: when the two alleles that an individual carries are the same. For example, AA or aa.
- Heterozygous: when the two alleles that an individual carry are different. For example, Aa.
- Wild type: the "normal" allele or phenotype for an organism. The wild-type is usually the most prevalent, although it doesn't necessarily have to be true.
- Recessiveness: the "weak" allele. The recessive allele is only expressed if both copies are present. Only a single copy is needed for the dominant allele. The recessive allele is usually denoted as the lower case letter, the dominant allele is usually denoted as the upper case letter. For example, blond hair is recessive. Both alleles for blond hair need to be present, otherwise the hair is dark.
- Complete dominance
| Genotype | Phenotype |
| AA | Dominant |
| Aa | Dominant |
| aa | Recessive |
- Co-dominance
| Genotype | Phenotype |
| AA | A |
| AB | Both A and B |
| BB | B |
An example of co-dominance is the A and B blood type alleles. Type A cells have A antigens. Type B cells have B antigens. Type AB makes both antigens.
- Incomplete dominance, leakage, penetrance, expressivity
- Incomplete dominance:
| Genotype | Phenotype |
| AA | A |
| AB | In between A and B |
| BB | B |
An example of incomplete dominance is the color of chickens. A cross between black chickens and white chickens give rise to bluish grey chickens.
- leakage: gene flow from one species to another.
- Penetrance is the frequency that a genotype will show up in the phenotype. 100% penetrance means that if you have the genes for being smart, then you'll definitely be smart! Less than 100% penetrance means that you may have the genes for being smart, but you may not actually be smart.
- Expressivity is to what degree a penetrant gene is expressed. Constant expressivity means that if your genes for being smart manages to penetrate (show up as a trait), then you're IQ is 120. Variable expressivity means that your IQ doesn't have to be 120, it could be somewhat lower or somewhat higher.
- Hybridization: viability
- Hybrid vigor = increased viability for offspring of parents who are genetically more different
- Mechanism = less chance to receive 2 copies of the same detrimental recessive gene
- Gene pool: all of the alleles in a population.
Meiosis and genetic variability
- Significance of meiosis: meiosis introduces genetic variability by genetic recombination. Genetic recombination is the product of independent assortment and crossing-over, which introduces genetic variability.
- Important differences between meiosis and mitosis
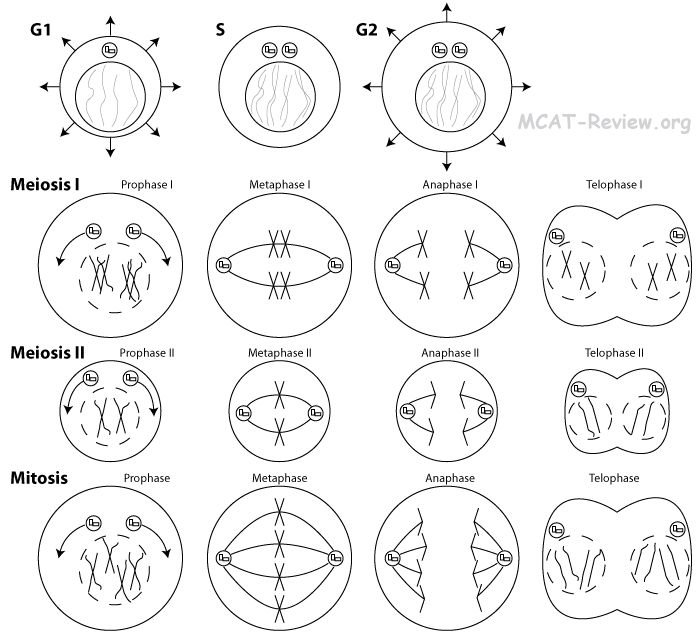
| mitosis |
meiosis |
| no tetrad |
tetrad formation (pairing of homologous chromosomes) and cross over |
| daughter cells identical to parent cell |
daughter cells different from parent cell |
| diploid (2n) daughter cells |
haploid (n) daughter cells |
| 1 division involved |
2 divisions involved |
| 2 daughter cells |
4 sperm cells or 1 egg (with polar bodies) |
- Segregation of genes
- Independent assortment
- Independent assortment generates genetic variation.
- A cell has 2 copies of each somatic chromosome- one from mom, one from dad (homologous chromosomes). Independent assortment shuffles these chromosomes, and then places only one copy of each into the gamete. This way, the gamete may have chromosome 1 from mom, chromosome 2 from dad, chromosome 3 from dad, ... etc.
- The mechanism of independent assortment is the following: During metaphase I of meiosis, homologous chromosome pair up along the metaphase line in random orientation - sometimes the mom's chromosome is on the left, sometimes it's on the right. During anaphase I of meiosis, the homologous chromosomes are pulled apart. Those on the left will be put into one daughter cell, those on the right will be put into another.
- Linkage
- Because of independent assortment, genes on different chromosomes are randomized. However, genes on the same chromosome can not be randomized by this mechanism.
- Genes on the same chromosome are linked to some extent.
- Crossing over is a mechanism that reduces linkage. However, crossing over is only efficient when the genes are physically apart from each other on the chromosome.
- When the genes are further apart on the chromosome, crossing over makes them less linked.
- The physically closer the genes are on the chromosome, the more linked they are.
- Recombination: also called genetic recombination, is the process that introduces genetic diversity into the gametes during meiosis. There are 2 processes that makeup recombination: independent assortment and crossing over.
- Crossing over occurs during prophase I (the actual site of cross over is the chiasma. The chiasma is made possible because of pairing of homologous chromosomes called the tetrad, which is formed by a process called synapsis).
- Single crossovers: results in genetic recombination. The chromatids involved in this single crossover exchange alleles at a given locus. Results in 2/4 recombinants.
- Double crossovers:
- Scenario 1: results in no genetic recombination. The chromatids involved in this double crossover exchange alleles at first, but then it exchanges them back, resulting in no net recombination. This is called the 2-strand double crossover. Results in 0/4 recombinants.
- Scenario 2: results in genetic recombination. The chromatids exchange alleles during a crossover. Then, one of the crossover chromatid exchanges with a different chromatid. This is called the 3-strand double crossover. Results in 2/4 recombinants.
- Scenario 3: results in genetic recombination. The chromatids exchange, then 2 totally different chromatids on the same chromosome exchange. This is called the 4-strand double crossover. Results in 4/4 recombinants.
- Synaptnemal complex: the protein complex that glues the tetrad together
- Tetrad: the paired homologous chromosome structure
- Sex-linked characteristics = gene for the characteristic is on the X chromosome.
- Very few genes on Y chromosome
- The Y chromosome is very small and carries few genes of importance.
- All the sex-linked alleles are carried on the X chromosome.
- Sex determination: XX = female, XY = male
- Cytoplasmic/extranuclear inheritance
- Cytoplasmic inheritance = inheritance of things other than genomic DNA.
- All cellular organelles, such as mitochondria, is inherited from the mother.
- Mutation
- General concept of mutation-error in DNA sequence
- Mutation = change in DNA sequence by means other than recombination.
- Types of mutations: random, translation error, transcription error, base substitution, inversion, addition, deletion, translocation, mispairing
- Random mutation = random changes in DNA sequence. Can be due to radiation, chemicals, replication error ...etc.
- Translation error = even if the DNA for a gene is perfect, errors during translation can cause expression of a mutant phenotype.
- Transcription error = even if the DNA of a gene is perfect, errors during transcription can cause expression of a mutant phenotype.
- Base substitution = mutation involving a base (ATGC) changing to a different base.
- Inversion = a stretch of DNA (a segment of a chromosome) breaks off, then reattaches in the opposite orientation.
- Addition = also called insertion = an extra base is added/inserted into the DNA sequence.
- Deletion = a base is taken out of the DNA sequence.
- Addition/insertion and deletion mutations result in a frameshift mutation.
- Translocation = a stretch of DNA (a segment of a chromosome) breaks off, then reattaches somewhere else.
- Mispairing = A not pairing with T, or G not pairing with C.
- Advantageous vs. deleterious mutation
- Advantageous = results in a benefit to the fitness of the organism. For example, the mutation that causes flies to become wingless is advantageous in an environment that is very windy.
- Deleterious = results in a harmful effect to the fitness of the organism. For example, a mutation that causes an organism to be sterile.
- Inborn errors of metabolism = genetic diseases resulting in faulty metabolism. For example PKU (Phenylketonuria) is an inborn error of metabolism where people can't metabolize phenylalanine. There's no cure, but the treatment involves avoiding things containing the amino acid phenylalanine.
- Relationship of mutagens to carcinogens
- Mutagen = something that causes mutation.
- Carcinogen = something that causes a mutation that causes cancer.
- All carcinogens are mutagens.
- Not all mutagens are carcinogens.
- Genetic drift = random changes in allele frequency due to chance = not due to natural selection
- Synapsis or crossing-over mechanism for increasing genetic diversity
- Synapsis: homologous chromosomes come together and form tetrad
- Crossing-over: exchange of genetic material between homologous chromosomes after the tetrad forms
- Increases genetic diversity by introducing new allele combinations in the chromosome you're about to give to your offspring
Analytic methods
- Hardy-Weinberg Principle
- p+q = 1
- (p+q)2 = 1 → p2 + 2pq + q2 = 1
- Five Assumptions of Hardy-Weinberg
- Infinitely large population (no genetic drift)
- No mutation
- No migration
- Random mating (no sexual selection)
- No natural selection
- Test cross: back cross, concepts of parental, F1 and F2 generations
- Test cross: so you have something with dominant phenotype. It could either be Aa or AA. To find out, you cross it with the homozygous recessive aa. If Aa, half the offspring will express the recessive phenotype. If AA, no offspring will express the recessive phenotype.
- Back cross = mating between the offspring and the parent = preserve parental genotype.
- Parental generation = P = generation of the parent. On a pedigree, the is the row that represents the parents
- F1 generation = Felial 1 = children. On a pedigree, this is the row below the parents, and represents the children of the parents.
- F2 generation = Felial 2 = grandchildren. On a pedigree, this is the row below the F1, and represents the children of the F1 and grandchildren of the parents.
- Gene mapping: crossover frequencies
- Gene mapping = physical location genes on the chromosome (eg. toward the end, closer to the middle, etc)
- Further apart = higher crossover frequency
- Biometry: statistical methods
- Biometry = using statistics to analyze biological data
- null hypothesis = first assuming that you are wrong - there is no relationship in your data
- p value = calculated chance that the null hypothesis is right, that you are wrong
- p value < 0.05 means less than 5% chance the null hypothesis is right (basically the null hypothesis is wrong, you are right, there is a relationship in your data)
- t-test: compares 2 data sets
- ANOVA: compares 3+ data sets
- Fisher exact test: compares data in a 2x2 table
- Variance, standard deviation: evaluates the distribution of a single set of data. Higher value = data is more spread out
- Skew = asymmetry in the bell curve. Skewed left = longer "tail" of the bell curve on the left.
|
|
