|
|
Enzyme structure and function
- Function of enzymes in catalyzing biological reactions
- Enzymes are catalysts, which are things that increase the rate of a reaction, but does not get used up during the reaction.
- Structure determines function. A change in structure => a change in function.
- Important biological reactions catalyzed by enzymes:
- Metabolism
- DNA synthesis
- RNA synthesis
- Protein synthesis
- Digestion
- Enzyme classification by reaction type
- Enzyme class = reaction type + "ase"
- Oxidase, reductase = oxidation, reduction reaction
- Dehydrogenase = removes hydrogen (oxidation), can also work in reverse (reduction)
- Kinase (adds), phosphatase (removes) phosphate groups
- Protease, amylase, lipase = hydrolyzes proteins, carbohydrates, fats
- DNAse, RNAse, nuclease = hydrolyzes phosphodiester bonds in nucleic acids
- Polymerase = makes DNA/RNA/protein (polymers of nucleic acids/amino acids)
- Transcriptase = transcribes RNA from DNA template (reverse transcriptase makes DNA from RNA template)
- Ligase = forms bonds between 2 strands of DNA/RNA
- Reduction of activation energy
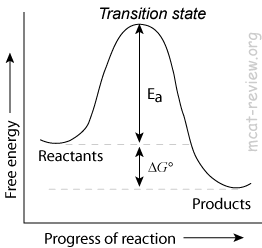 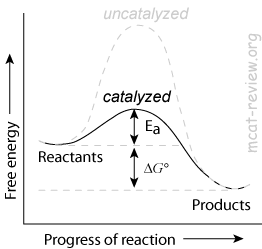
- Enzymes decrease the activation energy (Ea) of a reaction by lowering the energy of the transition state.
- Enzymes increase the rate of a reaction by decreasing the activation energy.
- Enzymes will increase the rate constant, k, for the equation rate = k[A][B].
- Enzymes do NOT change the Keq of a reaction.
- Enzymes do not change Keq because it lowers the activation energy for BOTH forward and reverse reactions.
- Enzymes will make the reverse reaction go faster also.
- Enzymes do not change ΔG, the net change in free energy.
- Enzymes affect the kinetics of a reaction, but not the thermodynamics.
- Substrates and enzyme specificity
- Enzyme-substrate interactions occur at the enzyme's active site.
- Enzyme-substrate specificity derives from structural interactions.
- Lock and key model: rigid active site. Substrate fits inside the rigid active site like a key.
- Induced fit model: flexible active site. Substrate fits inside the flexible active site, which is then induced to "grasp" the substrate in a better fit.
- Enzymes can be specific enough to distinguish between stereoisomers.
- Mechanisms of catalysis
- Enzymes can be protein or RNA.
- Almost all enzymes in your body is made of protein.
- The most important RNA enzyme in your body is the ribosome.
- Structure determines function; Enzyme structure derives from 4 levels.
- Primary: this is the sequence of the protein or RNA chain.
- Secondary: this is hydrogen bonding between the protein backbone. Examples include alpha helices and beta sheets (backbone H-bonding). For RNA, this is base pairing.
- Tertiary: this is the 3-D structure of the enzyme. This involves -R group interactions and spatial arrangement of secondary structure.
- Quaternary: when more than 1 chain is involved. When you hear about "dimers", "trimers", "tetramers", "oligomers", that's quaternary structure.
- Cofactors = metal ions (DNA polymerase needs magnesium)
- Coenzymes = small molecules (NAD, FAD, CoA, vitamins)
- Fat soluble vitamins: Vit A, D, E, K. Can't be excreted in urine, so can be toxic at high levels
- A: night vision (night blindness if deficient)
- D: bones (Rickets if deficient in kids, Osteomalacia in adults)
- E: cell membranes (deficiency causes hemolytic anemia, neuropathy, myopathy)
- K: clotting (Bleed if deficient. Warfarin blocks vitamin K pathway and works as a blood thinner. Premature infants lack vitamin K because they haven't had time to acquire the gut bacteria that makes vitamin K)
- Water soluble vitamins: anything that's not fat soluble. Less toxic as it can be peed out
- B1/thiamin: deficient in alcoholics (Wernicke-Korsakoff syndrome) or dietary deficiency (Beriberi)
- B12/cobalamine: needed in DNA synthesis. Acquired through diet. Deficient in strict vegetarians or inflammatory bowel disease. Causes macrocytic anemia and neuropathy
- Folate: needed in DNA synthesis. Deficient in alcoholics or dietary lack of vegetables. Causes macrocytic anemia. Anti-folate drugs block folate pathways to kill rapidly dividing (DNA synthesizing) cancer cells.
- Vitamin C: needed in collagen synthesis. Deficiency causes scurvy
- Effects of local conditions on enzyme activity
- Enzymes have optimal conditions (temperature, pH, salt concentration) where they function the best
- Usually this is body temperature, but the immune system functions better at 1 degrees higher. This is why you get a fever when you get an infection.
- Too much fever can denature enzymes and cause permanent damage. This is why antipyretic drugs (acetaminophen) are used to treat fever.
Control of enzyme activity
- Kinetics
- General (catalysis)
- Enzyme "catalyzes" a reaction, meaning it changes the kinetics of a reaction without being used up
- Kinetics (how fast a reaction occurs) vs Thermodynamics (whether and what direction a reaction will occur)
- Enzyme makes a reaction go faster by lowering the activation energy
- Enzymes lower the activation energy by stablizing the transition state (high energy state between reactant and product)
- Michaelis-Menten = reaction rate (V) vs substrate concentration [S] plot
- Vmax = maximum reaction rate = V where [S] is infinity
- Km = Michaelis constant = [S] where V is half of Vmax
- Cooperativity
- Positive cooperativity = binding of substrate from one subunit makes another subunit more likely to bind
- Negative cooperativity = binding of substrate from one subunit makes another subunit less likely to bind
- Feedback regulation
- The product of a pathway inhibits the pathway.
- For example, hexokinase, the first enzyme in glycolysis, is inhibited by its product glucose-6-phosphate.
- Inhibition - types
- Competitive inhibition
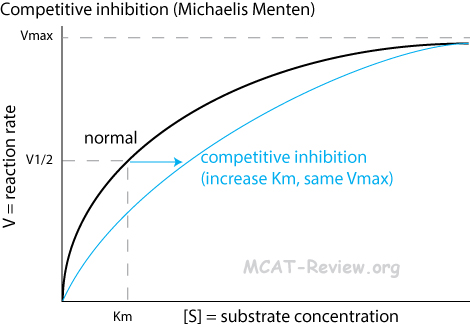
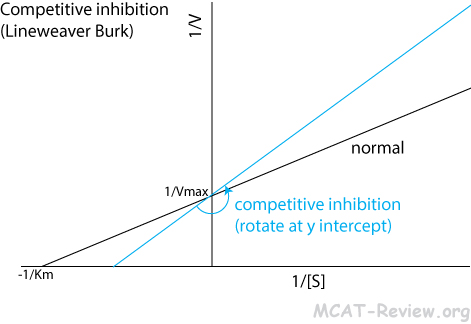
- An inhibitor competes with the substrate for binding to the active site.
- Competitive inhibition increases Km (the amount of substrate needed to achieve maximum rate of catalysis).
- Competitive inhibition does NOT change Vmax (the maximum possible rate of the enzyme's catalysis).
- Michaelis Menten plot: less hyperbolic curve
- Lineweaver Burk plot: counterclockwise rotation around the y intercept
- You can overcome competitive inhibition by providing more substrate.
- Non-competitive inhibition
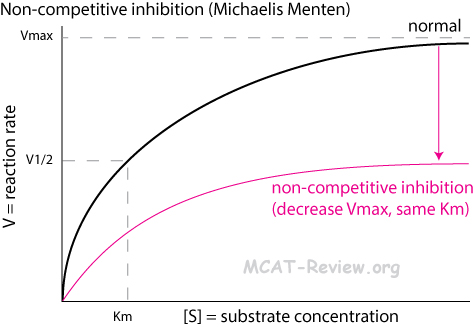
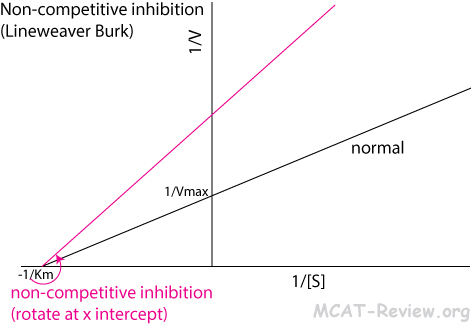
- An inhibitor binds to an allosteric site on the enzyme to deactivate it.
- The substrate still have access the active site, but the enzyme is no longer able to catalyze the reaction as long as the inhibitor remains bound.
- Non-competitive inhibition decreases Vmax (the maximum possible rate of the enzyme's catalysis).
- Non-competitive inhibition does NOT change Km (the amount of substrate needed to achieve the maximum rate of catalysis).
- Michaelis Menten plot: lower plateau
- Lineweaver Burk: counterclockwise rotation around the x intercept
- You can't overcome non-competitive inhibition by adding more substrate.
- Mixed
- Michaelis Menten plot: lower plateau AND less hyperbolic curve
- Lineweaver Burk plot: both y and x intercept changes (y intercept shifts up, x intercept shifts right)
- Uncompetitive inhibition
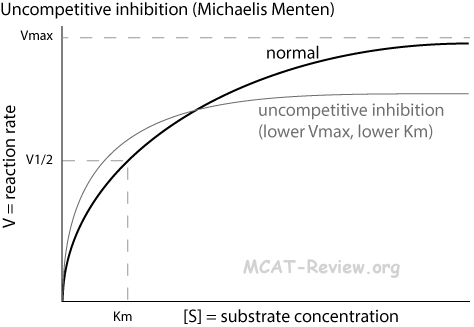
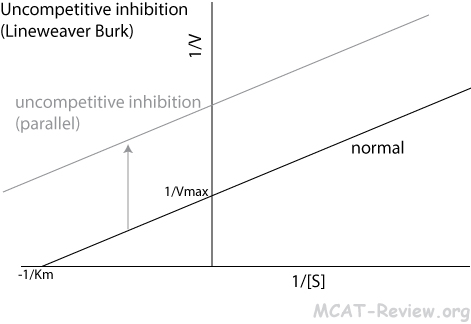
- Inhibitor binds the enzyme-substrate complex
- Michaelis Menten plot: lower plateau but more hyperbolic curve
- Lineweaver Burk plot: parallel shift upward
- Regulatory enzymes
- Allosteric enzymes: binding at allosteric site -> change in enzyme conformation -> change in active site to bind substrate more easily
- Covalently-modified enzymes: phosphorylation of an enzyme makes it active/inactive
- Zymogen: inactive until activated by another enzyme. Eg: Pepsinogen (inactive) -> pepsin (active); trypsinogen (inactive) -> trypsin (active)
|
|
